SemiNMF-PCA framework for Sparse Data Co-clustering
Allab, Labiod, and Nadif (2016) in CIKM
Research Paper
Christian Bager Bach Houmann @ AAU
9 Jan 2024
The problem
- Given a large set of documents and their content, how can we group them into topics and understand which terms are typically associated?
- Existing methods struggle with high-dimensionality and sparsity in document-term matrices, doesn’t capture the underlying geometry well
- Most clustering approaches only focus on documents or terms, but not both (unilateral clustering)
- Using PCA and then k-means clustering (Tandem clustering) is discouraged
Proposed solution: Integrating SemiNMF and PCA
Understanding the SemiNMF-PCA Framework for Sparse Data Co-clustering
- Semi non-negative matrix factorization (SemiNMF)
- Principal Component Analysis (PCA) for
- Sparse data
- Co-clustering
Sparse, High-Dimensional Data
- Sparse Data: Dataset where most entries are zero
- High Dimensionality: Datasets with a large number of features (e.g., terms in text dataset)
- Key Challenges:
- Difficulty in visualizing and understanding the structure
- Traditional clustering methods fall short in capturing the underlying patterns
Brief introduction to NMF and PCA
- Nonnegative Matrix Factorization (NMF):
- Decomposes data matrices into parts for easier interpretation
- Data must be non-negative
- Principal Component Analysis (PCA):
- Reduces dimensions while keeping the most important variability
Co-clustering
- What is Co-clustering?
- Simultaneous grouping of data points (e.g., documents) and features (e.g., terms).
- Why Co-clustering?
- Often more effective than unilateral clustering, especially for sparse, high-dimensional data
- With matrix factorization
- NMF for unilateral, then NMTF for co-clustering
Co-clustering
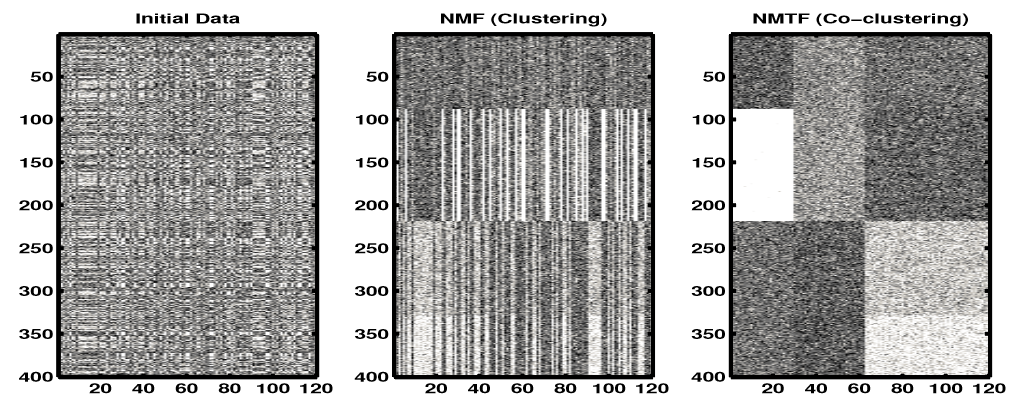
Clustering vs. Co-clustering: Allab, Labiod, and Nadif (2016)
Matrix factorization based co-clustering algorithms
- Croeuc Algorithm: Uses the principle of double k-means to perform co-clustering on continuous data
- Bipartite Spectral Graph Partitioning Algorithm (Spec): Designed to be sequel to Croeuc
- Information-Theoretic Co-clustering (ITCC)
- Spectral Co-clustering Algorithm (SpecCo): Appears to perform well on document clustering
Locality Preserving based Co-clustering
- Drawback of other methods: they often overlook local manifold geometry because they mainly rely on global Euclidean geometry
- Dual Regularized Co-Clustering (DRCC): Combines NMTF with incorporating manifold structures in both sample and feature spaces. Limited in handling data with negative values and has high computational complexity.
- Locally Preserved Fast Nonnegative Matrix Tri-Factorization (LpFNMTF): Tries to reduce computational demands by enforcing constraints for factors to be cluster indicator matrices
SemiNMF
- Semi-nonnegative:
- data \(X\) and cluster centroids \(S\) can be positive and negative, but
- cluster indicator matrix \(G\) must be zero or positive: a data point has a non-negative degree of association with a cluster
- Soft clustering: a point can have partial membership to a cluster
- Goal is to minimize difference between \(X\) and \(GS^{\top}\)
PCA
- Used to find lower-dimensional subspace best representing the data
- Done by identifying principal directions \(Q\) and projecting the data points into this new subspace, giving principal components \(U\)
- Goal is reconstructing \(UQ^{\top}\) as closely to \(X\) as possible, subject to \(Q^{\top}Q=I\) (ensures orthogonality)
- \(U\) can be considered continuous analogue to the discrete membership indicators in k-means
- This is the basis for Laplacian embedding integration
SemiNMF-PCA-CoClust
Form matrix \(M\) of size \((n+d)\times(n+d)\) from data \(X\): \[ M=\left[\begin{matrix} 0 & X \\ X^{\top} & 0 \end{matrix}\right] \]
- Obtain \(G\) by doing k-means clustering on \(X\) and \(X^{\top}\) - represents cluster membership for both samples and features
- \(S\) consists of centroid matrices for samples and features
- \(Q\) contains components reduced from SVD of \(X\)
Goal is minimizing: \[ \min_{G,S,Q}\| M-GSQ^{\top} \|^{2}\quad s.t.\quad G\geq 0, Q^{\top}Q=I \]
Regularized SemiNMF-PCA CoClust
- Uses graph Laplacian embeddings to account for intrinsic geometric structure
- Create KNN data graph for samples and features: data points connected to k nearest neighbors
- Used to represent local proximity/similarity between points, capturing local structure and relationship in data
- Create weight matrices \(W_g\) and \(W_f\) from graph: represents connections in data samples and features, respectively
- Compute normalized graph Laplacians \(L_g\) and \(L_f\) from \(W_g, W_f\) and diagonal matrices of them
Regularized SemiNMF-PCA CoClust
Introduce the normalized graph Laplacians in \(M\): \[ M=\left[\begin{matrix} \alpha L_{g} & X \\ X^{\top} & \beta L_{f} \end{matrix}\right] \] where \(\alpha\) and \(\beta\) are the regularization parameters used to control the contribution of \(L_{g}\) and \(L_{f}\) respectively.
The minimization problem is of the same form as before, but with updated \(M\).
\[ \min_{G,S,Q}\| M-GSQ^{\top} \|^{2}\quad s.t.\quad G\geq 0, Q^{\top}Q=I \]
Optimize by updating \(S\), \(G\), and then \(Q\) repeatedly until convergence.
Experimental Results and Analysis
Performance Metrics
- Accuracy (Acc): correctness of cluster asignments
- Normalized Mutual Information (NMI): evaluate mutual information between cluster assignments and true labels
- Adjusted Rand Index (ARI): Assess similarity between clusterings and ground truth, adjusting for chance
Data and Parameters
- Datasets size and sparsity varied across datasets:
- Samples (documents): min=~500, max=~20k
- Features (terms): min=~1k, max=~43k
- Run each method w. different parameter settings 50x
- Reports best results per metod
- Grid search regularization parameter \(\alpha\) with \(\beta=\alpha0.1\) in grid \((0.01, 0.1, 1, 10, 100, 500, 1000)\)
Results
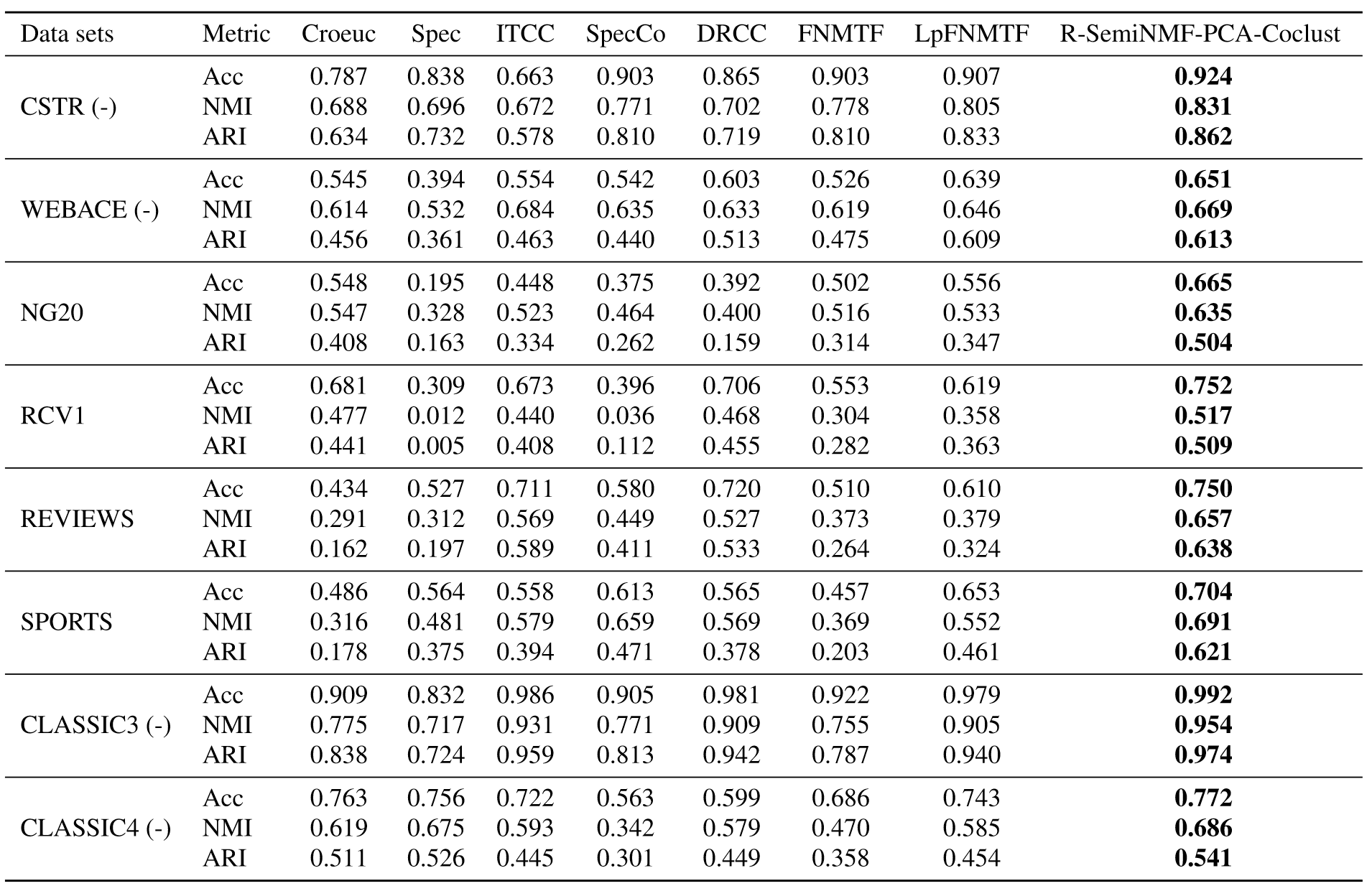
Allab, Labiod, and Nadif (2016)
Results: Statistical Tests
- One-way ANOVA & pairwise t-tests
- Show statistically significant performance increase over LpFNMTF
Cluster Visualization

Allab, Labiod, and Nadif (2016)
- The method groups clusters to provide clearer separation between them; there’s less overlap
Regularization parameters
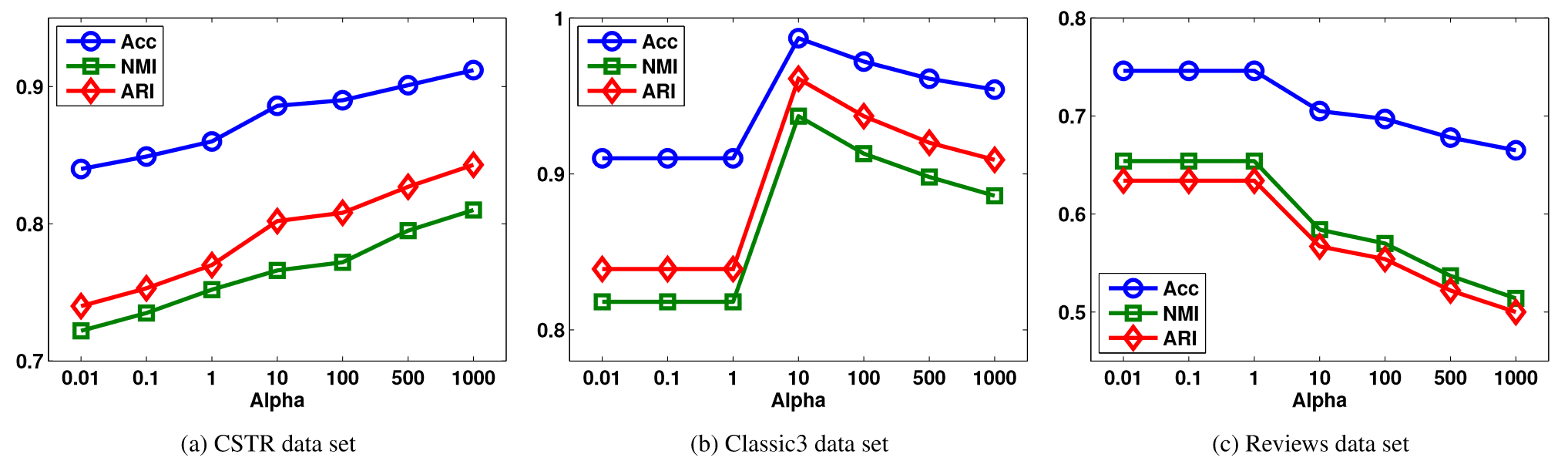
Allab, Labiod, and Nadif (2016)
- CSTR: Performance increases with increase in \(\alpha\)
- Classic3: Optimal at \(\alpha=10\)
- Reviews: Performance decreases with increase in \(\alpha\)
- Choosing optimal parameters for datasets vs. generalization
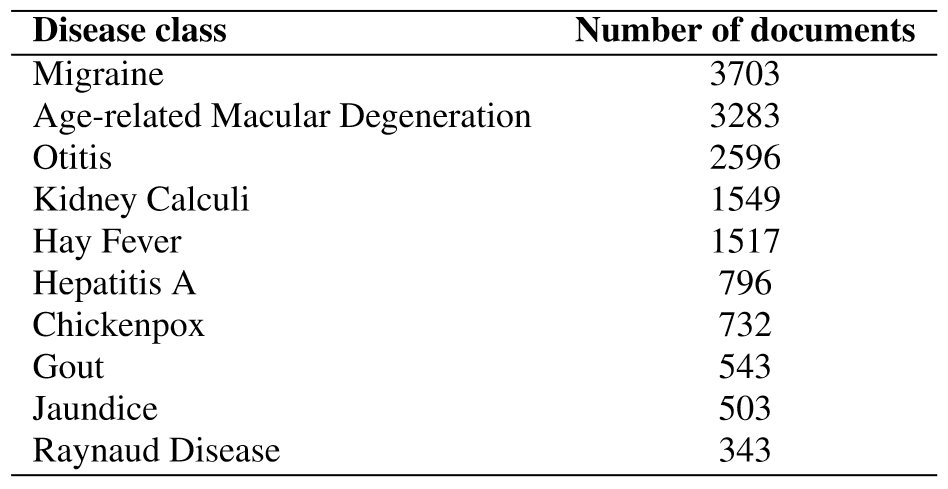
PUBMED
To illustate co-clustering capabilities on term clusters
PUBMED10
- Biomedical abstracts categorized by disease
- Divided into
- PUBMED10
- PUBMED6
- PUBMED5
Results

Allab, Labiod, and Nadif (2016)
Dense bands of variables - terms cited in many docs - considered noise.
Results
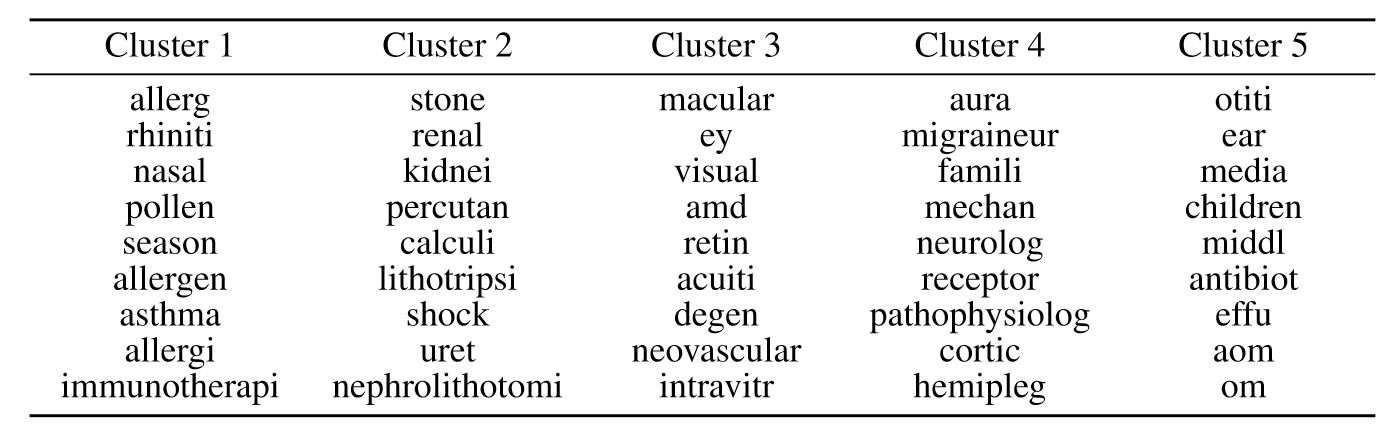
Allab, Labiod, and Nadif (2016)
- Identified semantically coherent column clusters indicative of document clusters - seems to correspond to diseases
- Found common terms between different diseases - showing ability to handle overlap and shared features between clusters
Conclusion
- Presented framework unifying dimensionality reduction and co-clustering
- Achieves better performance than previous methods
Critical Evaluation
Critical Evaluation: Limitations
- Motivation is lacking
- Questionable choices for mathematical terms
- Clear weakness that they didn’t evaluate co-clustering better; but they excuse it with there being no gold standard labels for terms.
- Unclear whether the method needs parameter tuning specific to the dataset1, but it somewhat seems that way
Critical Evaluation: Strengths
- Figures and illustrations were useful
- The frameworks seems to manage noise well1
- Integration is well reasoned and grounded in theory
- Achieved statistically significant increase in performance over state of the art
Critical Evaluation: Additional points
- Would have liked to see performance over all runs, not just via variance (std dev).
- Computational complexity is not described; how does it compare to other SoTA algorithms?
Q&A
- Thank you for your attention!
- I am now ready to answer your questions and discuss further.